- December 04, 2018
- By Matthew E. Wright And Samantha Watters
The International Union for Conservation of Nature’s (IUCN) Red List of Threatened Species is a powerful tool for researchers and policymakers working to stem the global tide of species loss. But adding even a single species demands countless hours of expensive, rigorous research. The result is that vast numbers of species have never been assessed.
A new method co-developed by Anahí Espíndola, an assistant professor of entomology, uses the power of a branch of artificial intelligence known as machine learning, along with open-access data, to help streamline the process. The method uses algorithms to predict plant species that could be eligible for at-risk status on the IUCN Red List.
Currently, only about 5 percent of all known plants appear in any capacity on IUCN’s Red List, which ranks species in five levels, from those of least concern to those that are critically endangered.
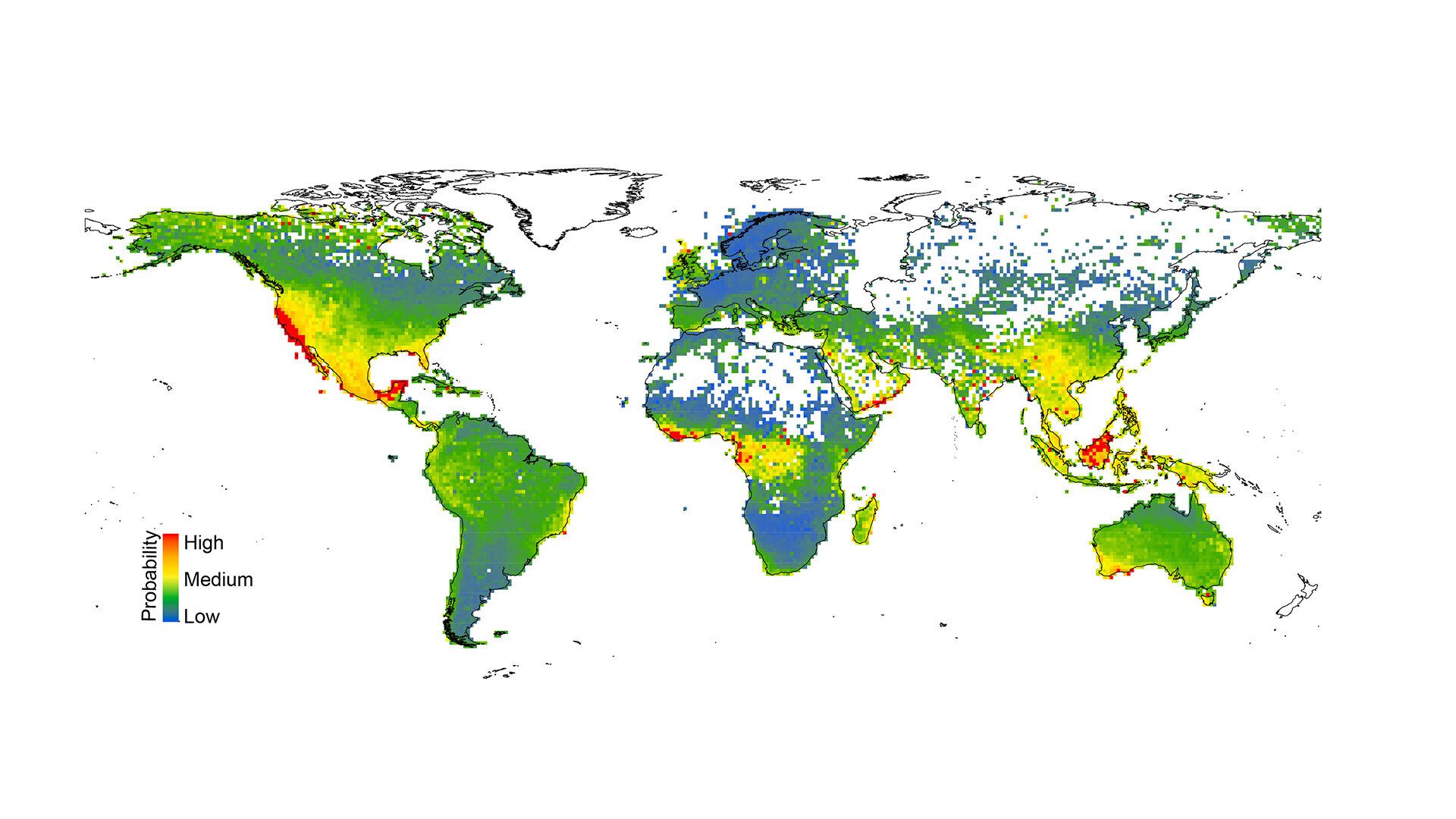
After training an algorithm to assess more than 150,000 plant species worldwide—one of the largest assessments of conservation risk yet conducted—Espindola and her colleagues found that more than 10 percent of the species they reviewed are highly likely to qualify for at-risk classification. The researchers published their findings online yesterday in the Proceedings of the National Academy of Sciences.
“Our method isn’t meant to replace formal assessments using IUCN protocols,” Espíndola said. “It’s a tool that can help prioritize the process, by calculating the probability that a given species is at risk. Ultimately, we hope it will help governments and resource managers decide where to devote their limited resources for conservation.”
She and her collaborators built their predictive model using open-access data from the Global Biodiversity Information Facility and the TRY Plant Trait Database. Lead author Tara Pelletier, an assistant professor of biology at Radford University, worked with Espíndola to perform the machine learning analysis.
Espíndola and Pelletier trained the model using data from the relatively small group of plant species already on the IUCN Red List. This allowed them to assess and fine-tune the model’s accuracy by checking its predictions against the listed species’ known IUCN risk status, before applying it to a broader range of species.
The researchers noted several major geographical trends in the model’s predictions: At-risk species tended to cluster in areas known for high native biodiversity, such as the Central American rainforests and southwestern Australia. The model also flagged regions such as California and the southeastern United States, which are home to a large number of endemic species, meaning that they do not naturally occur anywhere else on Earth.
The model also highlighted risks in a few surprising areas not typically known for their biodiversity, such as the southern coast of the Arabian Peninsula, as having a high number of at-risk species. Some of the most imperiled regions have not received enough attention from researchers, according to Espíndola. She hopes that her method can help to fill in some of these knowledge gaps by identifying regions and species in need of further study.
“Everything we’ve done is 100 percent open access, highlighting the power of publicly available data,” Espíndola said. “We hope people will use our model—and we hope they point out errors and help us fix them, to make it better.”